Nature Chemistry April 2018: Abstract
Robert Alberstein, Yuta Suzuki, Francesco Paesani, and F. Akif Tezcan
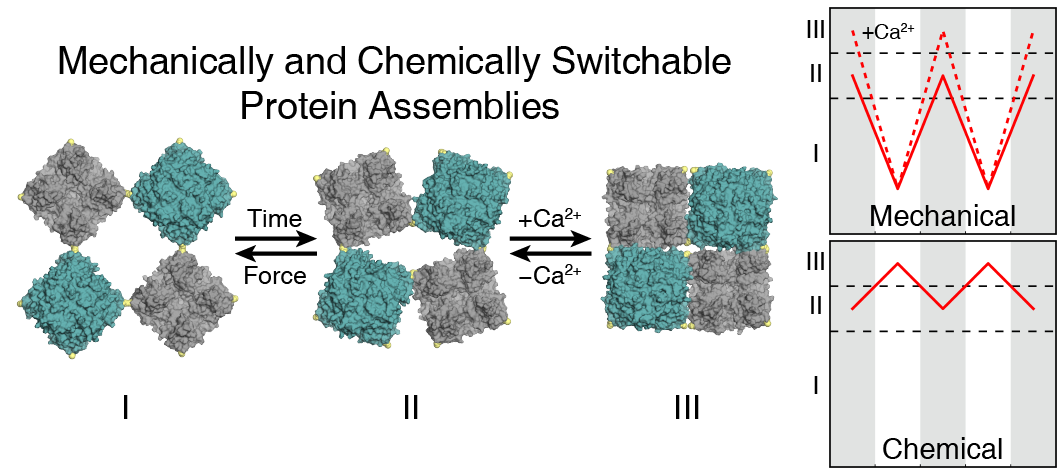
- De novo design and construction of stimuli-responsive protein assemblies that predictably switch between discrete conformational states remains an essential but highly challenging goal in biomolecular design. We previously reported synthetic, two-dimensional protein lattices self-assembled via disulfide bonding interactions, which endows them with a unique capacity to undergo coherent conformational changes without losing crystalline order. Here, we carried out all-atom molecular dynamics simulations to map the free-energy landscape of these lattices, validated this landscape through extensive structural characterization by electron microscopy and established that it is predominantly governed by solvent reorganization entropy. Subsequent redesign of the protein surface with conditionally repulsive electrostatic interactions enabled us to predictably perturb the free-energy landscape and obtain a new protein lattice whose conformational dynamics can be chemically and mechanically toggled between three different states with varying porosities and molecular densities.
Read the Behind the Paper post by Robert Alberstein here!
Check out the writeup and podcast by the Texas Advanced Computing Center covering this work here!